We want to highlight three publications that have utilized CRCF instruments and acknowledged RI-INBRE Funding. The topics include DNA repair, asymmetric synthesis, and repair of DNA adducts.
Biocore T200 Surface Plasmon Resonance (SPR):Comparative Studies on Bulky DNA Damage Binding by Nucleotide Excision Repair Proteins Using Surface Plasmon Resonance, Differential Scanning Fluorometry, and DNase I Footprinting. Chemical Research Toxicology, 2024, 38, p206. This paper was featured as a Journal Cover.
Circular Dichroism (CD), Bruker Avance DRX 400 MHz spectrometer: Asymmetric synthesis of an atropisomeric β-carboline via regioselective intermolecular Rh(I)-catalyzed [2 + 2 + 2] cyclotrimerization. Tetrahedron Letters 2024, 146, p155187.
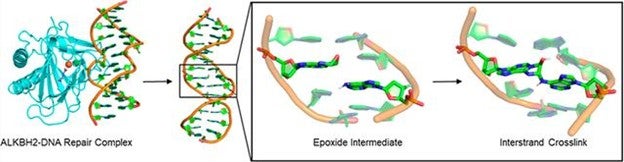
AB Sciex 4600 Mass Spectrometry: Stable Interstrand Cross-Links Generated from the Repair of 1,N6-Ethenoadenine in DNA by α-Ketoglutarate/Fe(II)-Dependent Dioxygenase ALKBH2. Journal of American Chemical Society 2024, 146, p10381.
Cai, A.; LaVigne, K. L.; Crisalli, A. M.; Delaney, S.; Min, J.-H.; Cho, B. P. Comparative studies on bulky DNA damage binding by nucleotide excision repair proteins using surface plasmon resonance, differential scanning fluorometry, and DNase I footprinting. Chemical Research in Toxicology 2025, 38 (1), 206–215. DOI: https://doi.org/10.1021/acs.chemrestox.4c00456.
 This paper highlights a highly collaborative work among colleagues from three universities, providing critical insights into how the repair protein XPC recognizes and interacts with bulky DNA lesions. Led by the RI-INBRE Core Director Ang Cai under the guidance of the RI-INBRE Program Director Bongsup Cho (URI) with key contributions from Dr. Sarah Delaney and Katelyn L. LaVigne at Brown University, and Junghyun Min at Balor University, the study investigates how the human XPC protein and its yeast ortholog Rad4 interact with bulky di-FAAF-modified DNA lesions. Using advanced CRCF items—surface plasmon resonance (SPR)/Biacore T200 instrument and differential scanning fluorimetry (DSF)/Tycho NT.6 system, as well as DNase I footprinting—the team revealed that XPC binds damaged DNA with 10-fold greater affinity and 16-fold stronger retention than Rad4, while Rad4 undergoes more significant conformational changes upon binding. DNase I footprinting identified a 7-nucleotide protected region on the complementary strand, showing Rad4’s interaction with the undamaged DNA. This collaborative work, leveraging cutting-edge instrumentation from CRCF, provides critical insights into DNA damage recognition and repair mechanisms, with implications for cancer research and therapeutic development.
This paper highlights a highly collaborative work among colleagues from three universities, providing critical insights into how the repair protein XPC recognizes and interacts with bulky DNA lesions. Led by the RI-INBRE Core Director Ang Cai under the guidance of the RI-INBRE Program Director Bongsup Cho (URI) with key contributions from Dr. Sarah Delaney and Katelyn L. LaVigne at Brown University, and Junghyun Min at Balor University, the study investigates how the human XPC protein and its yeast ortholog Rad4 interact with bulky di-FAAF-modified DNA lesions. Using advanced CRCF items—surface plasmon resonance (SPR)/Biacore T200 instrument and differential scanning fluorimetry (DSF)/Tycho NT.6 system, as well as DNase I footprinting—the team revealed that XPC binds damaged DNA with 10-fold greater affinity and 16-fold stronger retention than Rad4, while Rad4 undergoes more significant conformational changes upon binding. DNase I footprinting identified a 7-nucleotide protected region on the complementary strand, showing Rad4’s interaction with the undamaged DNA. This collaborative work, leveraging cutting-edge instrumentation from CRCF, provides critical insights into DNA damage recognition and repair mechanisms, with implications for cancer research and therapeutic development.
Hughes, R. R.; Battistoni, L. D.; Ciesla, M. J.; Bolton, T. j.; Asher, P. M.; Irizarry, G.; de Jesus Antonio Martinez, A.; Baker, K. M.; Mulcahy, S. P. Asymmetric synthesis of an atropisomeric β-carboline via regioselective intermolecular Rh(I)-catalyzed [2 + 2 + 2] cyclotrimerization. Tetrahedron Letters 2024, 146, 155187. DOI: https://doi.org/10.1016/j.tetlet.2024.155187.
 Riley R. Hughes, Circular Dichroism (CD) spectroscopy was used to analyze the enantiomers 10-entA and 10-entB at the Central Research Core Facility (CRCF), providing crucial insights into their chiral characteristics
Riley R. Hughes, Circular Dichroism (CD) spectroscopy was used to analyze the enantiomers 10-entA and 10-entB at the Central Research Core Facility (CRCF), providing crucial insights into their chiral characteristics
Riley R. Hughes, an undergraduate student at Providence College, served as the first author of a publication co-authored with his mentor, Dr. Seann P. Mulcahy, a member of the RI-INBRE Executive Committee. Their paper, titled “Asymmetric synthesis of an atropisomeric β-carboline via regioselective intermolecular Rh(I)-catalyzed [2 + 2 + 2] cyclotrimerization,” presents a novel and efficient method for synthesizing atropisomeric β-carbolines—molecules with unique axial chirality that hold significant value in drug discovery, asymmetric catalysis, and chiroptical applications. The researchers developed a six-step synthetic strategy, including most notably a Grignard addition/elimination reaction to generate an yne-ynamide precursor, followed by a Rh(I)-catalyzed [2 + 2 + 2] cyclotrimerization with ethyl cyanoformate. This method yielded the target compounds with excellent efficiency, enantioselectivity, and regioselectivity. The team further enhanced the process’s effectiveness by optimizing key reaction conditions, such as solvent selection, catalyst/ligand combinations, and temperature.
The researchers employed advanced spectroscopic techniques to confirm the synthesized atropisomers’ structural integrity and chiral properties. Circular Dichroism (CD) spectroscopy was used to analyze the enantiomers 10-entA and 10-entB at the Central Research Core Facility (CRCF), providing crucial insights into their chiral characteristics. Additionally, Proton (1H) and Carbon (13C) Nuclear Magnetic Resonance (NMR) spectroscopy was conducted using a Bruker Avance DRX 400 MHz spectrometer at Providence College’s Chemistry Department, an extension laboratory of the RI-INBRE CRCF. These analyses confirmed the molecular structure and integrity of the synthesized compounds. Combining these techniques ensured the accuracy and reliability of the synthesis, laying a strong foundation for future developments in atropisomeric compounds with promising pharmaceutical and biological applications.
Wang J, Takyi NA, Hsiao YC, Tang Q, Chen YT, Liu CW, Ma J, Qi R, Bian K, Peng Z, Essigmann JM, Lu K, Wetmore SD, Li D. Stable Interstrand Cross-Links Generated from the Repair of 1,N6-Ethenoadenine in DNA by α-Ketoglutarate/Fe(II)-Dependent Dioxygenase ALKBH2. J Am Chem Soc. 2024 Apr 17;146(15):10381-10392. doi: 10.1021/jacs.3c12890. Epub 2024 Apr 4. PMID: 38573229; PMCID: PMC11060877.
 This paper, published in the Journal of the American Chemical Society, discovered a new type of DNA damage that happens when cells try to fix a specific DNA problem called 1,N6N6-ethenoadenine (eAeA), which is caused by things like inflammation or exposure to harmful chemicals. The damage, called an interstrand cross-link (ICL), occurs when a repair enzyme called ALKBH2 accidentally creates a strong bond between the damaged DNA and a healthy part of the opposite DNA strand. This bond can block critical cell processes like copying or reading DNA, which could lead to cell death or disease. The researchers used sophisticated instrumentation at CRCF such as AB Sciex 4600 mass spectrometry, ultra centrifuges, and computer simulations to show how this happens and even found evidence in human cells. This discovery helps us understand how DNA repair can sometimes go wrong and cause more harm, especially in conditions like cancer or after exposure to toxins.
This paper, published in the Journal of the American Chemical Society, discovered a new type of DNA damage that happens when cells try to fix a specific DNA problem called 1,N6N6-ethenoadenine (eAeA), which is caused by things like inflammation or exposure to harmful chemicals. The damage, called an interstrand cross-link (ICL), occurs when a repair enzyme called ALKBH2 accidentally creates a strong bond between the damaged DNA and a healthy part of the opposite DNA strand. This bond can block critical cell processes like copying or reading DNA, which could lead to cell death or disease. The researchers used sophisticated instrumentation at CRCF such as AB Sciex 4600 mass spectrometry, ultra centrifuges, and computer simulations to show how this happens and even found evidence in human cells. This discovery helps us understand how DNA repair can sometimes go wrong and cause more harm, especially in conditions like cancer or after exposure to toxins.