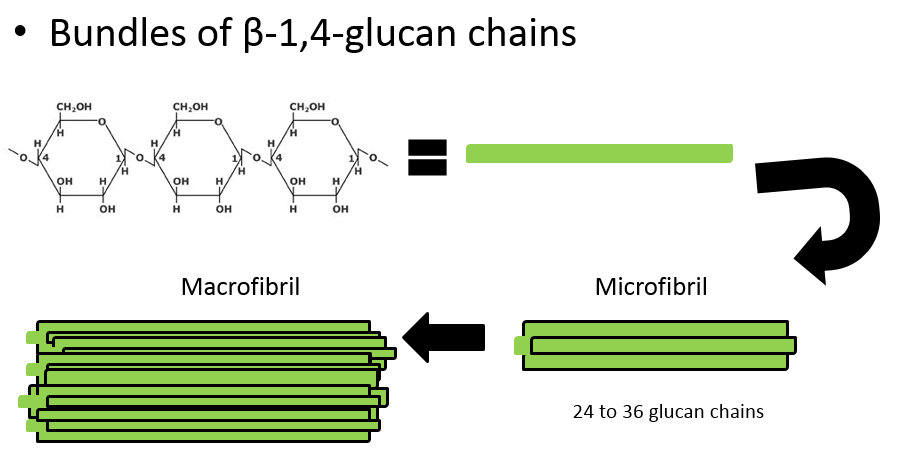
In the Roberts lab, we study plant cell walls. One of the main components of plant cell walls are cellulose microfibrils, long threads made up of multiple (usually 36) strands of glucose molecules linked together by ß(1-4) linkages. The threads are cross-linked by hydrogen bonds.
Additional important molecules such as pectins, glycoproteins, and lignins give different cell walls their particular characteristics and functions.
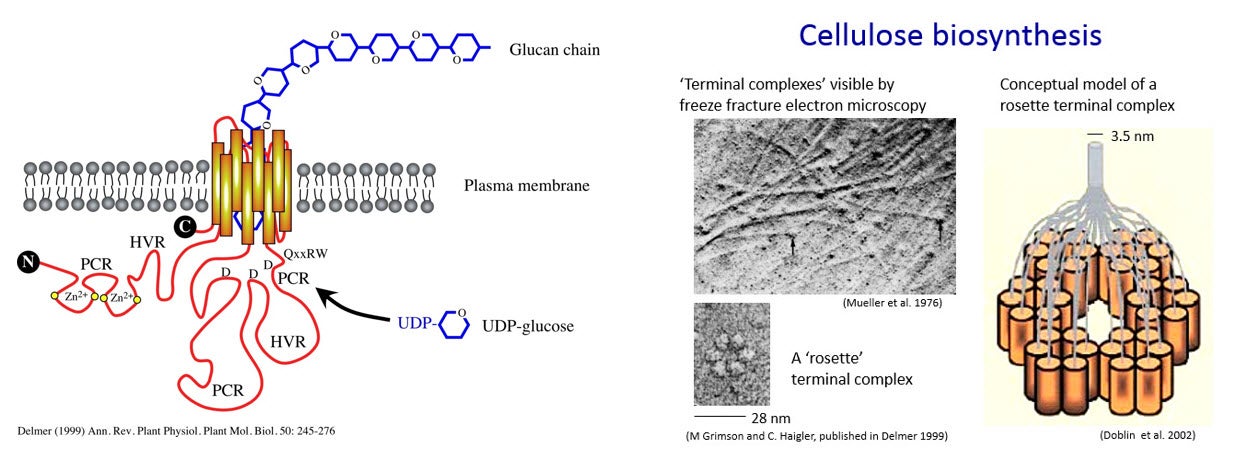
Cellulose microfibrils are synthesized by proteins, called CESAs, embedded in and moving transversely in the plasma membrane. These proteins are composed of multiple subunits arranged together in specific patterns, and are referred to as “terminal complexes”. In this lab, we investigate the genes that code for the proteins which when embedded in the plasma membrane produce cellulose microfibrils to the outside of the membrane.
The following animation shows how the crystallization of cellulose microfibrils helps to propel the terminal complex within the membrane (http://www.sciencedirect.com/science/article/pii/S000634950771072X):
microfibril synthesis animation
In comparing the genes that do this job in different organisms, we are learning about how the process of cellulose synthesis evolved. We are also investigating the arrangement of the protein subunits that form the terminal complexes.
The terminal complexes in different types of organisms take various shapes and result in the production of cellulose strands and ribbons of various shapes. For example, the red alga Porphyra yezoensis has terminal complexes arranged in a line, and produces flat ribbons of cellulose. In higher plants, the terminal complexes are arranged in a hexagonal rosette as seen in the figure above. This arrangment produces cylindrical cellulose microfibrils approximately 3.5 x 3.5 nm in cross section (Delmer 1999).
How do we study the genes for CESAs?
CESA genes have already been sequenced from higher plants such as Arabidopsis (the fruit fly of plants), rice and poplar, algae such as Mesotaenium, and prokaryotes such as Acetobacter. Some regions of the genes are the same in all of these, while other regions differ. These regions that are the same are helpful in finding new genes in other organisms.
Using some sequences from those areas of the genes that all CesAs seem to have in common, primers are created for use in PCR to find complementary sequences in cDNA from other species. Once these other genes are found, we can sequence them, study when and where they are expressed, and try to knock out their function and analyze the resulting phenotype in an attempt to deduce their role in the plant.
Plasmids called “knockout vectors” are created that ….. In our case, the knockout vector contains upstream and downstream DNA sequence of the targeted gene and in the middle, it has antibiotic resistant cassette. When the DNA is introduced into the moss, homologous recombination occurs upstream and downstream of the target gene. The target gene is then replaced by the antibiotic selection cassette, thus knocking out the gene of interest.
Transformation
Once we have a knock out vector constructed, we have to get the DNA into the plant cells. The following is a summary of the protocol that we use to accomplish this transformation.
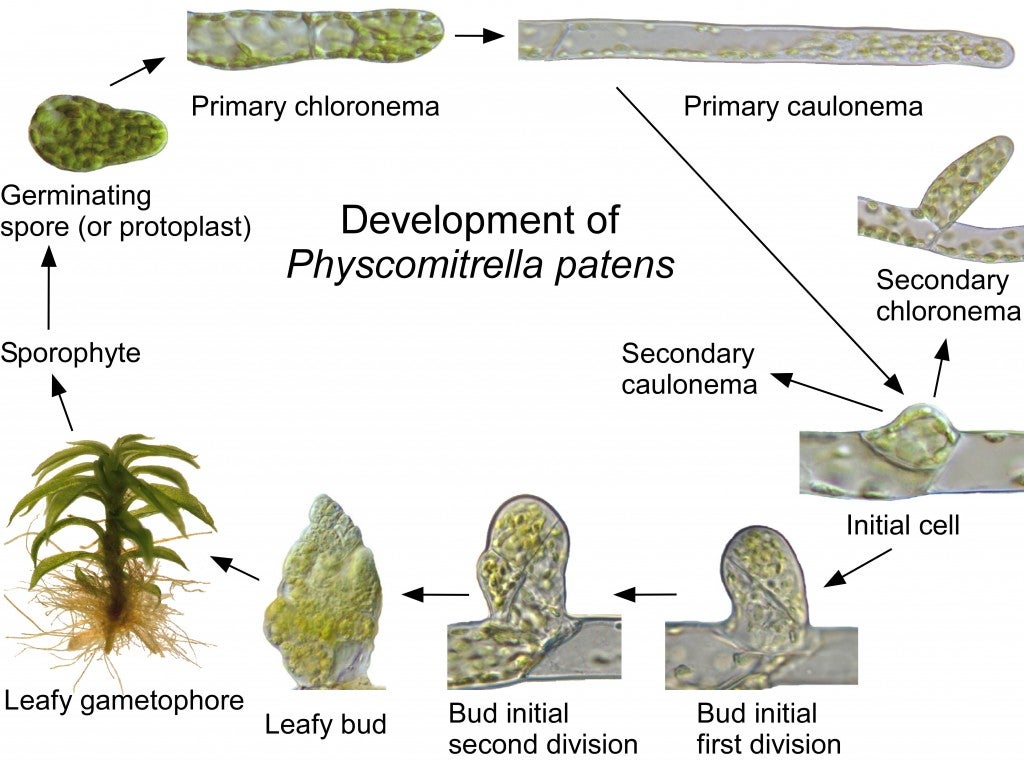
The moss cultures (Physcomitrella patens) must be grown under sterile conditions in Petri dishes so that there will be no contaminating bacteria or fungi. All of the moss from one plate is scraped from one culture dish and incubated with an enzyme called Driselase that will break down the cell walls of the moss protonemata cells and release the individual cells. These “protoplast” cells no longer have cell walls, and could burst due to water entering the cell by osmosis, so this step is carried out in a solution of mannitol, a sugar alcohol.
The protoplasts are pelleted in a centrifuge to remove the cell debris, rinsed with fresh mannitol solution, and then incubated with the DNA molecules for transformation. The lightning bolt in the figure is a “heat-shock” step in which the movement of the DNA into the cells is accelerated by 3 minutes at 45 degrees C. It is not really understood how the DNA molecules get across the plasma membrane, and then into the nucleus, but it works in at least some of the protoplasts!
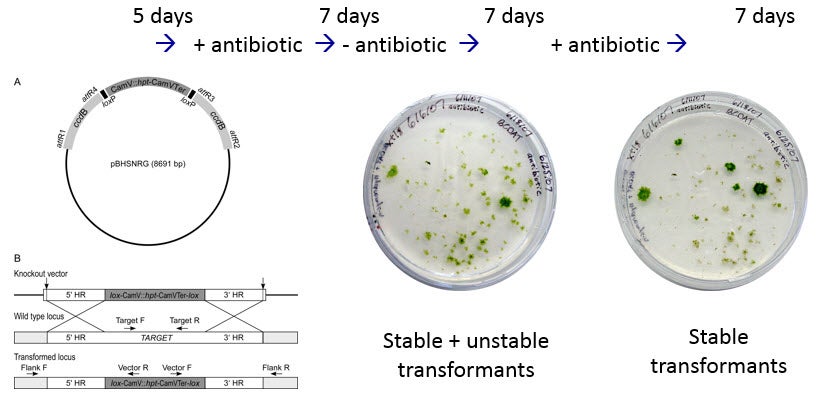
Now the protoplasts are spread thinly on a culture plate containing mannitol, and allowed to grow and divide to form a new protonemata filament for 5 days. Then they are transferred to a culture plate containing the antibiotic hygromyocin. This is easy because the protoplasts are growing on a cellophane circle on the culture dish! After 7 days, the protoplasts that did not take up the DNA molecules containing the hygromyocin resistance gene will die (black dots).
Then the protoplasts are transferred to media without antibiotic for another week, and then transferred back to culture plates containing hygromyocin for the final week. This should kill any colonies in which the DNA entered the cell, but did not cross over into the chromosomes. This is called a transient transformation because eventually the DNA molecules would be lost, and the cells would no longer be resistant to hygromyocin. The larger black dots in the last image represent the dead transient colonies on the second hygromyocin plate. The green colonies are stable transformants in which the DNA molecule has been incorporated into the chromosome. But only some of these clones will have knocked out the gene with a true cross-over event. Others will have simply inserted the DNA into the chromosome without knocking out the gene. Genotyping by PCR must be done to determine which colonies are true knock-outs.
Works Cited
Delmer 1999
Roberts and Roberts 2009
 Home
Home Browse
Browse Close
Close Events
Events Maps
Maps Email
Email Brightspace
Brightspace eCampus
eCampus