Multi-institution team uses quantitative imaging technology, innovative robotic device to capture tissue in minutes, preserve for advanced genomic study
Jan. 17, 2024
A University of Rhode Island professor of Ocean Engineering and Oceanography, along with a multidisciplinary research team from multiple institutions, successfully demonstrated new technologies that can obtain preserved tissue and high-resolution 3D images within minutes of encountering some of the most fragile animals in the deep ocean.
Professor Brennan Phillips (GSO Ph.D. ’16), the principal investigator on the project, and a team of 15 researchers from six institutions, including URI, have shown that it is possible to shave years from the process of determining whether a new or rare species has been discovered. The results of their work are published today in the journal Science Advances.
Roboticists, ocean engineers, bioengineers, and marine and molecular biologists from URI’s Department of Ocean Engineering; the Bigelow Laboratory for Ocean Sciences in East Boothbay, Maine; the School of Engineering and Applied Sciences at Harvard University; Monterey Bay Aquarium Research Institute (MBARI) in California; PA Consulting, a worldwide firm that focuses on innovation; and the Department of Natural Sciences at Baruch College, City University of New York, made up the team. The paper represents five years of research.

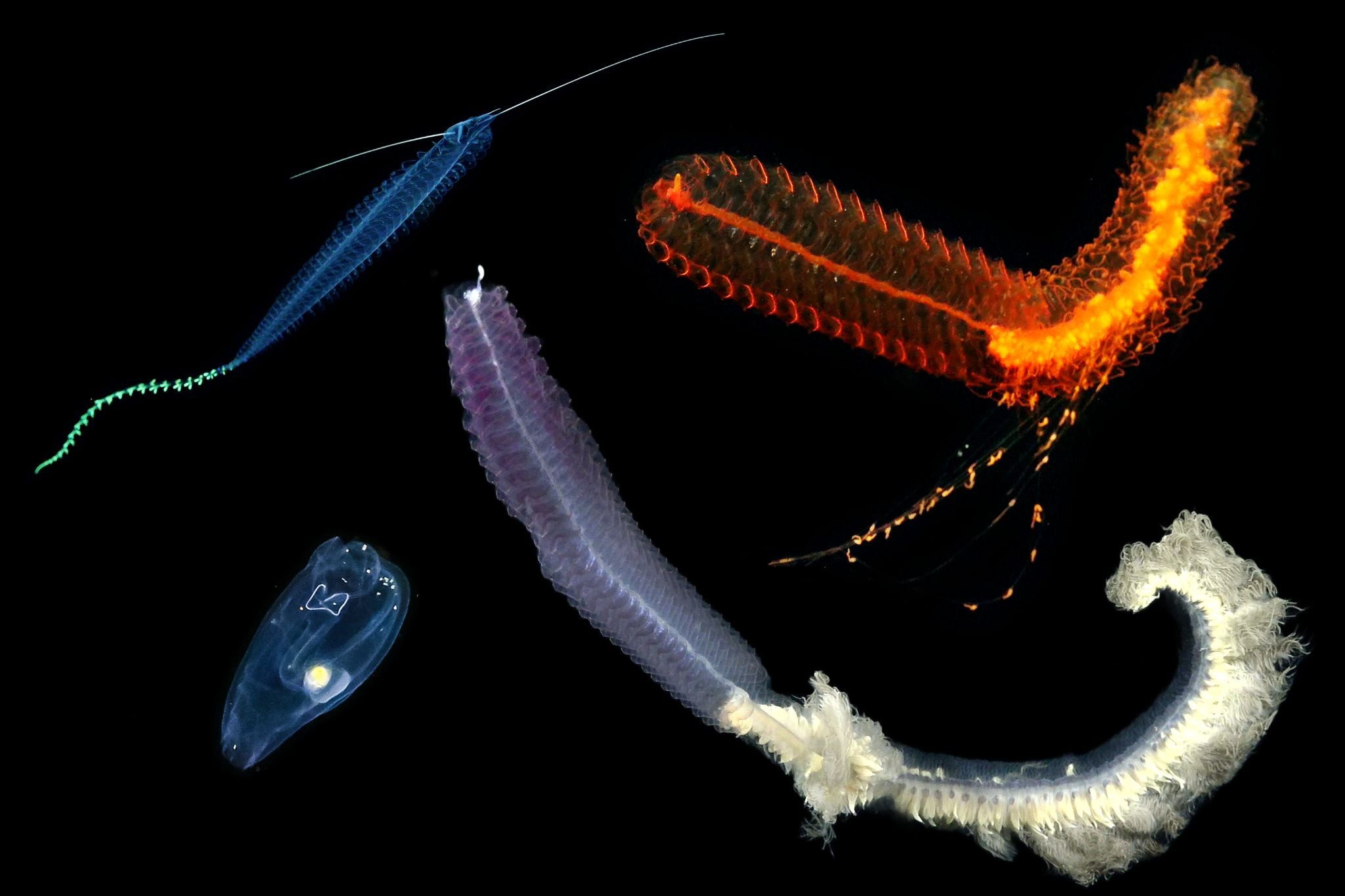
Revolutionary advancements in underwater imaging, robotics, and genomic sequencing have reshaped marine exploration, the study reads. The research shows that within minutes of an encounter with a deep-sea animal, it is possible to capture detailed measurements and motion of the animal, obtain an entire genome, and generate a comprehensive list of genes being expressed that point to their physiological status in the deep ocean. The result of this rich digital data is a ‘cybertype’ of a single animal, rather than a physical ‘holotype’ that is traditionally found in museum collections.
“Currently, if researchers want to describe what they believe is a new species, they face an arduous process,” Phillips said. “The way it is done now is you capture a specimen, which is very difficult because a lot of these animals are so delicate and tissue-thin, and it’s likely you may not be able to collect them at all. But if you successfully collect an animal, you then preserve it in a jar. Then begins a long process of physically bringing that specimen to different collections around the world where it is compared to existing organisms. After a long time, sometimes up to 21 years, scientists may reach consensus that this is a new species.
“Again, these are deep-sea, thin little wisps of animals. The current workflow is not appropriate. It’s a major reason why we have so many undescribed species in the ocean.”

Information gained from the study—and others that follow—could be useful for extinction prevention studies, as it provides a wealth of information from a single specimen gained during a single encounter. The work also responds to the growing call among researchers for compassionate collection, which minimizes harm to animals by using advanced technologies to collect information. Future studies and development could allow for complete scans and inventories of life in the deep sea within a catch-and-release framework.
“The vision was: How might a marine biologist work to better understand and connect to deep-sea life decades or centuries into the future?” said David Gruber, Distinguished Professor of Biology at Baruch College, City University of New York, and an Explorer with National Geographic Society. “This is a demonstration on how an interdisciplinary team could work collaboratively to provide an enormous amount of new information on deep-sea life after one brief encounter. The ultimate goal is to continue down this path and refine the technology to be as minimally-invasive as possible—akin to a doctor’s check-up in the deep sea! This approach is becoming increasingly important with current extinction rates being 100 times higher than background extinction rates.”

Phillips said because collecting these samples has always been hard, there are many deep-sea species that have yet to be identified. “When you look at climate change and deep-sea mining and their potential effects, it is unsettling,” Phillips said. “You realize you don’t have a full baseline of species, and you may not know what you’ve lost before it’s too late. If you want to know what has been there before it’s gone, this is a new way to do that.”
The mission, which was funded by the Schmidt Ocean Institute and its Designing the Future program, and conducted on its research vessel Falkor, included two expeditions off the coast of Hawaii and San Diego in 2019 and 2021. The team collected as many as 14 preserved tissue samples a day, along with terabytes of quantitative digital imagery. Together, the study provided:
- The first complete assembled and annotated transcriptome (genes being made in the animals’ habitat) ofPegea tunicate, a marine invertebrate animal;
- Details of the molecular basis of environmental sensing of a holoplanktonic Tomopteris polychaete (marine worm), which spends its entire life in the water column;
- Details of the full transcriptomes of two siphonophores, (gelatinous zooplankton composed of specialized parts growing together in a chain) Erenna sp. and Marrus claudanielis, as well as the Pegea tunicate and Tomopteris polychaete;
- Full morphological (form and structure) characterizations using digital imaging of each animal while at depth.
The lead author of the paper, John Burns, a senior research scientist at Bigelow Laboratory, conducted the genomic analysis on four animals sampled at depths of almost 4,000 feet.
“What we were able to achieve with these animals is remarkable,” Burns said. “For me, this is best seen in the sequence data we generated for the Tomopteris worm: We captured it while it was exploring its environment and were able to infer that it was scanning the water using two long sensory whiskers near its head for ‘sweet’ tastes: likely sugars associated with prey, and possibly for ammonia: a waste product of its typical prey.
“With that information, we can envision how it hunts by following chemical trails in its open water habitat,” Burns said. “I don’t think that would have been possible without the innovative technology invented and employed by the engineers on the team that allowed complete preservation of the information from the animals within minutes of an encounter.”
Burns said another study with Gruber looked at how capture methods affect jellyfish ribonucleic acid, known as RNA, one of the building blocks of life. That sequence of information can start to change after about 10 minutes of stressful conditions, even with gentle collection. The Designing the Future technologies overcome this by preserving the information before the animal’s cells start to respond to stress, according to Burns.
“We also discovered that three of the animals we captured have huge genomes: each having nearly 10 times the DNA in a cell compared to us humans!” Burns said. “For the fourth, with a more modestly sized genome (about 3% the size of a human genome) we were able to use cutting edge sequencing methods to build the most cohesive and complete genome of a salp to date.”
Harvard and URI brought to the mission a rotary-actuated folding dodecahedron (RAD-2), an innovative origami-inspired robotic encapsulation device, which collected animal tissue samples and almost instantaneously preserved that tissue at depth.
“We are seeing the impact of new types of marine robots for midwater and deep-sea exploration,” said roboticist Robert Wood, the Harry Lewis and Marlyn McGrath Professor of Engineering and Applied Sciences at Harvard University. “Not only are robots going places that are difficult or impossible for humans to reach, our devices investigate, interact with, and collect specimens using a gentle touch… or no touch at all.”
Imaging systems from MBARI’s Bioinspiration Lab that included a laser-scanning imaging device called DeepPIV and a three-dimensional lightfield camera called EyeRIS enabled the measurement and reconstruction of three-dimensional morphology, or body shape, of the animals in their natural environment.
“We cannot protect what we do not yet fully understand. Advanced imaging technologies can accelerate our efforts to document the diversity of life in the ocean. The faster we can catalog marine life, the better we can assess and track the impact of human actions like climate change and mining on ocean environments,” said Kakani Katija, bioengineer and principal engineer of the Bioinspiration Lab at MBARI.
“We have these remotely operated vehicles out there with advanced imaging systems, which can create a three-dimensional model after only a few minutes,” Phillips said. “We were able to approach a tiny jellyfish in a matter of seconds, collect high-resolution 3D images to the control room, and our team was able to tell in a matter of minutes that the tentacles were exactly 5 millimeters long. Then, we had extremely well-preserved tissue samples of the same animal within a matter of minutes.”