November 22, 2021
Like the bacteria in your gut, single-celled marine plants, diatoms, harbor a diverse microbiome. Diatoms are responsible for about 20% of the world’s oxygen and their microbiome helps them grow. In a new paper, recent URI Graduate School of Oceanography graduate Olivia Ahern (Ph.D. ’21) and several colleagues found that the diatom’s bacterial microbiome composition depends on the host diatom’s genetics.
This collaborative research, funded by the Gordon and Betty Moore Foundation and RI NSF EPSCoR, was published on November 22nd in the journal Proceedings of the National Academy of Sciences.
Ahern’s discovery is the latest step in a research career sparked more than a decade ago during a formative experience at the Graduate School of Oceanography. Ahern, the new paper’s first author and a Newport, Rhode Island native, first experienced the wonders of oceanography as a high school intern for Leslie Smith (Ph.D. ‘11), a then-Ph.D. student in Professor Candace Oviatt’s lab on the URI Bay Campus. College plans took Ahern to South Carolina where she attended the College of Charleston and studied marine biology with a minor in studio art. Unable to resist being away from New England for long, Ahern moved back to Rhode Island upon graduation and spent time working for the Narragansett Bay Estuary Program.
Ahern was eager to learn more about oceanography, a subject that initially piqued her interest. Luckily, GSO was close by. She decided to take a couple of core oceanography classes as a non-matriculated student, during which she realized how fascinating a career in oceanography could be. “After meeting many people during my internship, I knew I wanted to come back to GSO for graduate school,” said Ahern.
While spending time on campus getting to know people and the lay of the land, Ahern met with GSO professor Tatiana Rynearson about potential projects in her lab. When asked about her transition to graduate school, Ahern said, “Even though I didn’t know much about molecular techniques or diatoms at the time, I joined the Rynearson Lab and was ready to learn all that I could.”
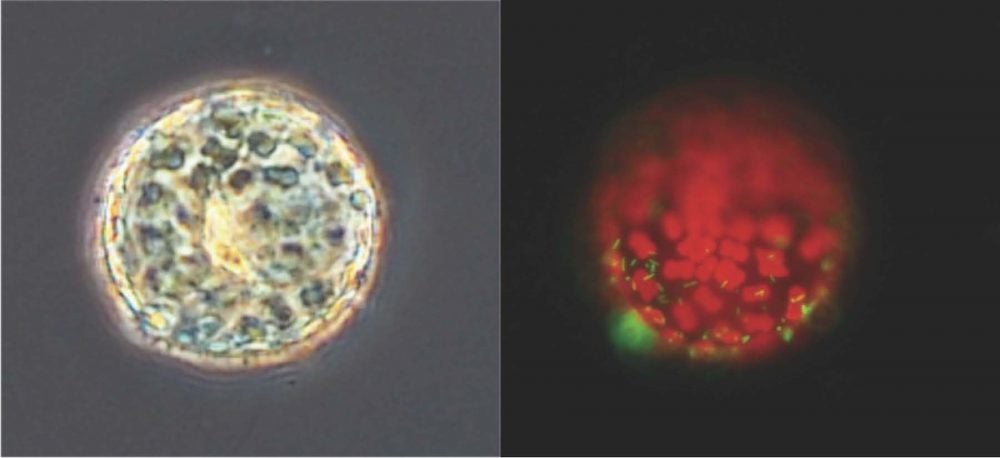
Once she started doing research in the lab, Ahern worked on diatom microbiomes. That is, she investigated interactions at the interface between diatom cells, a type of phytoplankton, and the bacteria that surround these cells. Just like we harbor unique bacteria on our skin or in our gut, diatoms harbor bacteria within their microbiome. After many hours in the lab and behind a computer screen, Ahern transferred into the Ph.D. program to continue to probe this secret life of diatoms.
Ahern specifically remembers the point at which the idea for her recent paper was born. During one of their weekly meetings, she and Rynearson examined some data and discussed Ahern’s observations. “As I saw that the patterns of microbiome composition were similar to those of diatom populations, I wanted to investigate the underlying cause for this,” Ahern said.
Ahern was intrigued because she wondered what could cause these similarly observed patterns. Was it that the diatom host genetics and microbiome are related?
Research comes with its challenges, and as Ahern said, “One of the hardest parts of this work was to prove this potential relationship between diatom and bacteria populations.”
At first, she used metagenomics, the collective set of genes from bacteria in her environmental samples. However, she could not obtain enough DNA for this approach.
Eager to persevere and see the project through to completion, Ahern and her co-authors of the paper published on November 22nd characterized the bacterial microbiome of the diatom Thalassiosira rotula. For this work, Ahern and her co-authors used eight genetically distinct populations of T. rotula that GSO alum Kerry Whittaker (Ph.D. ’14) isolated. The diatom populations were not separated by geographic boundaries, meaning that one population of T. rotula can coexist simultaneously in different ocean basins. Ahern and her co-authors found that T. rotula had no core microbiome, meaning not one bacterial species was found across all the diatom microbiomes. They also found that microbiome composition was not influenced by environmental factors. Instead, diatoms of the same population but from different ocean basins had the same bacteria in their microbiome.

These findings indicate that the diatom’s microbiome composition depended on the host diatom’s genetics. They confirmed this observation with a common garden experiment in the lab. This experiment involved growing sterile, or bacteria-free, T. rotula cultures from different populations and spiking these cultures with the same bacterial community. The results of this experiment confirmed that diatom genetics influences the microbiome and “broadened the scope of this work beyond a biogeographical survey,” said Ahern.
Researchers have found links between host genetics in their microbiomes in terrestrial life, including humans and plants. In large land plants, these microbiomes predetermined by host genetics can influence growth rate, protect against disease and influence ecosystem-wide processes such as carbon flow. “Diatoms represent one of the smallest organisms to host microbiomes and the role of the diatom’s genotype, or genetic material, on structuring bacterial interactions was virtually unknown prior to our research,” said Ahern. “So, this is a pretty exciting finding.”

These microscale interactions have global implications, as diatoms are the base of the marine food web and rely on bacteria for vitamins and nutrients.
Through perseverance, Ahern and her co-authors uncovered exciting research findings that have implications for the ocean ecosystem. After being involved in such a significant contribution to the scientific community, Ahern is keen to continue her quest of answering ecologically important questions.
With her Ph.D. dissertation complete, Ahern is now a postdoctoral scientist under supervisors Joseph Vallino (Marine Biological Laboratory) and Julie Huber (Woods Hole Oceanographic Institution) where she uses a combination of approaches to study the microbial food web. In this position, she actively builds upon her skillset of both laboratory techniques and computer-based techniques for DNA analysis.
While reflecting on her time at GSO, Ahern says she appreciates the skills and knowledge she gained throughout her time there, from a high school intern to a doctoral graduate.
– Written by Diana Fontaine, GSO Ph.D. candidate
